You can run this notebook in a live session 
Dask DataFrames¶
We finished Chapter 1 by building a parallel dataframe computation over a directory of CSV files using dask.delayed. In this section we use dask.dataframe to automatically build similiar computations, for the common case of tabular computations. Dask dataframes look and feel like Pandas dataframes but they run on the same infrastructure that powers dask.delayed.
In this notebook we use the same airline data as before, but now rather than write for-loops we let dask.dataframe construct our computations for us. The dask.dataframe.read_csv function can take a globstring like "data/nycflights/*.csv" and build parallel computations on all of our data at once.
When to use dask.dataframe¶
Pandas is great for tabular datasets that fit in memory. Dask becomes useful when the dataset you want to analyze is larger than your machine’s RAM. The demo dataset we’re working with is only about 200MB, so that you can download it in a reasonable time, but dask.dataframe will scale to datasets much larger than memory.

The dask.dataframe module implements a blocked parallel DataFrame object that mimics a large subset of the Pandas DataFrame API. One Dask DataFrame is comprised of many in-memory pandas DataFrames separated along the index. One operation on a Dask DataFrame triggers many pandas operations on the constituent pandas DataFrames in a way that is mindful of potential parallelism and memory constraints.
Related Documentation
Main Take-aways
Dask DataFrame should be familiar to Pandas users
The partitioning of dataframes is important for efficient execution
Create data¶
[1]:
%run prep.py -d flights
Setup¶
[2]:
from dask.distributed import Client
client = Client(n_workers=4)
We create artifical data.
[3]:
from prep import accounts_csvs
accounts_csvs()
import os
import dask
filename = os.path.join('data', 'accounts.*.csv')
filename
[3]:
'data/accounts.*.csv'
Filename includes a glob pattern *, so all files in the path matching that pattern will be read into the same Dask DataFrame.
[4]:
import dask.dataframe as dd
df = dd.read_csv(filename)
df.head()
[4]:
| id | names | amount | |
|---|---|---|---|
| 0 | 15 | Hannah | -954 |
| 1 | 66 | Jerry | 347 |
| 2 | 53 | Charlie | 4169 |
| 3 | 76 | Kevin | 2012 |
| 4 | 96 | Patricia | 2063 |
[5]:
# load and count number of rows
len(df)
[5]:
30000
What happened here? - Dask investigated the input path and found that there are three matching files - a set of jobs was intelligently created for each chunk - one per original CSV file in this case - each file was loaded into a pandas dataframe, had len() applied to it - the subtotals were combined to give you the final grand total.
Real Data¶
Lets try this with an extract of flights in the USA across several years. This data is specific to flights out of the three airports in the New York City area.
[6]:
df = dd.read_csv(os.path.join('data', 'nycflights', '*.csv'),
parse_dates={'Date': [0, 1, 2]})
Notice that the respresentation of the dataframe object contains no data - Dask has just done enough to read the start of the first file, and infer the column names and dtypes.
[7]:
df
[7]:
| Date | DayOfWeek | DepTime | CRSDepTime | ArrTime | CRSArrTime | UniqueCarrier | FlightNum | TailNum | ActualElapsedTime | CRSElapsedTime | AirTime | ArrDelay | DepDelay | Origin | Dest | Distance | TaxiIn | TaxiOut | Cancelled | Diverted | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| npartitions=10 | |||||||||||||||||||||
| datetime64[ns] | int64 | float64 | int64 | float64 | int64 | object | int64 | float64 | float64 | int64 | float64 | float64 | float64 | object | object | float64 | float64 | float64 | int64 | int64 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
We can view the start and end of the data
[8]:
df.head()
[8]:
| Date | DayOfWeek | DepTime | CRSDepTime | ArrTime | CRSArrTime | UniqueCarrier | FlightNum | TailNum | ActualElapsedTime | ... | AirTime | ArrDelay | DepDelay | Origin | Dest | Distance | TaxiIn | TaxiOut | Cancelled | Diverted | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1990-01-01 | 1 | 1621.0 | 1540 | 1747.0 | 1701 | US | 33 | NaN | 86.0 | ... | NaN | 46.0 | 41.0 | EWR | PIT | 319.0 | NaN | NaN | 0 | 0 |
| 1 | 1990-01-02 | 2 | 1547.0 | 1540 | 1700.0 | 1701 | US | 33 | NaN | 73.0 | ... | NaN | -1.0 | 7.0 | EWR | PIT | 319.0 | NaN | NaN | 0 | 0 |
| 2 | 1990-01-03 | 3 | 1546.0 | 1540 | 1710.0 | 1701 | US | 33 | NaN | 84.0 | ... | NaN | 9.0 | 6.0 | EWR | PIT | 319.0 | NaN | NaN | 0 | 0 |
| 3 | 1990-01-04 | 4 | 1542.0 | 1540 | 1710.0 | 1701 | US | 33 | NaN | 88.0 | ... | NaN | 9.0 | 2.0 | EWR | PIT | 319.0 | NaN | NaN | 0 | 0 |
| 4 | 1990-01-05 | 5 | 1549.0 | 1540 | 1706.0 | 1701 | US | 33 | NaN | 77.0 | ... | NaN | 5.0 | 9.0 | EWR | PIT | 319.0 | NaN | NaN | 0 | 0 |
5 rows × 21 columns
[9]:
df.tail() # this fails
distributed.worker - WARNING - Compute Failed
Function: execute_task
args: ((<methodcaller: tail>, (<function pandas_read_text at 0x7f9cb9177280>, <function read_csv at 0x7f9cba4e5700>, (<function read_block_from_file at 0x7f9cb9107160>, <OpenFile '/home/runner/work/dask-tutorial/dask-tutorial/data/nycflights/1999.csv'>, 0, 64000000, b'\n'), b'Year,Month,DayofMonth,DayOfWeek,DepTime,CRSDepTime,ArrTime,CRSArrTime,UniqueCarrier,FlightNum,TailNum,ActualElapsedTime,CRSElapsedTime,AirTime,ArrDelay,DepDelay,Origin,Dest,Distance,TaxiIn,TaxiOut,Cancelled,Diverted\n', {'parse_dates': {'Date': [0, 1, 2]}}, {'Date': dtype('<M8[ns]'), 'DayOfWeek': dtype('int64'), 'DepTime': dtype('float64'), 'CRSDepTime': dtype('int64'), 'ArrTime': dtype('float64'), 'CRSArrTime': dtype('int64'), 'UniqueCarrier': dtype('O'), 'FlightNum': dtype('int64'), 'TailNum': dtype('float64'), 'ActualElapsedTime': dtype('float64'), 'CRSElapsedTime': dtype('int64'), 'AirTime': dtype('float64'), 'ArrDelay': dtype('float64'), 'DepDelay': dtype('float64'), 'Origin': dtype('O'), 'Dest': dtype('O'), 'Dista
kwargs: {}
Exception: ValueError('Mismatched dtypes found in `pd.read_csv`/`pd.read_table`.\n\n+----------------+---------+----------+\n| Column | Found | Expected |\n+----------------+---------+----------+\n| CRSElapsedTime | float64 | int64 |\n| TailNum | object | float64 |\n+----------------+---------+----------+\n\nThe following columns also raised exceptions on conversion:\n\n- TailNum\n ValueError("could not convert string to float: \'N54711\'")\n\nUsually this is due to dask\'s dtype inference failing, and\n*may* be fixed by specifying dtypes manually by adding:\n\ndtype={\'CRSElapsedTime\': \'float64\',\n \'TailNum\': \'object\'}\n\nto the call to `read_csv`/`read_table`.')
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
/tmp/ipykernel_7074/668323394.py in <module>
----> 1 df.tail() # this fails
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/dask/dataframe/core.py in tail(self, n, compute)
1051
1052 if compute:
-> 1053 result = result.compute()
1054 return result
1055
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/dask/base.py in compute(self, **kwargs)
164 dask.base.compute
165 """
--> 166 (result,) = compute(self, traverse=False, **kwargs)
167 return result
168
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/dask/base.py in compute(*args, **kwargs)
442 postcomputes.append(x.__dask_postcompute__())
443
--> 444 results = schedule(dsk, keys, **kwargs)
445 return repack([f(r, *a) for r, (f, a) in zip(results, postcomputes)])
446
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/distributed/client.py in get(self, dsk, keys, restrictions, loose_restrictions, resources, sync, asynchronous, direct, retries, priority, fifo_timeout, actors, **kwargs)
2680 should_rejoin = False
2681 try:
-> 2682 results = self.gather(packed, asynchronous=asynchronous, direct=direct)
2683 finally:
2684 for f in futures.values():
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/distributed/client.py in gather(self, futures, errors, direct, asynchronous)
1974 else:
1975 local_worker = None
-> 1976 return self.sync(
1977 self._gather,
1978 futures,
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/distributed/client.py in sync(self, func, asynchronous, callback_timeout, *args, **kwargs)
829 return future
830 else:
--> 831 return sync(
832 self.loop, func, *args, callback_timeout=callback_timeout, **kwargs
833 )
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/distributed/utils.py in sync(loop, func, callback_timeout, *args, **kwargs)
337 if error[0]:
338 typ, exc, tb = error[0]
--> 339 raise exc.with_traceback(tb)
340 else:
341 return result[0]
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/distributed/utils.py in f()
321 if callback_timeout is not None:
322 future = asyncio.wait_for(future, callback_timeout)
--> 323 result[0] = yield future
324 except Exception as exc:
325 error[0] = sys.exc_info()
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/tornado/gen.py in run(self)
760
761 try:
--> 762 value = future.result()
763 except Exception:
764 exc_info = sys.exc_info()
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/distributed/client.py in _gather(self, futures, errors, direct, local_worker)
1839 exc = CancelledError(key)
1840 else:
-> 1841 raise exception.with_traceback(traceback)
1842 raise exc
1843 if errors == "skip":
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/dask/dataframe/io/csv.py in pandas_read_text()
149 df = reader(bio, **kwargs)
150 if dtypes:
--> 151 coerce_dtypes(df, dtypes)
152
153 if enforce and columns and (list(df.columns) != list(columns)):
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/dask/dataframe/io/csv.py in coerce_dtypes()
253 rule.join(filter(None, [dtype_msg, date_msg]))
254 )
--> 255 raise ValueError(msg)
256
257
ValueError: Mismatched dtypes found in `pd.read_csv`/`pd.read_table`.
+----------------+---------+----------+
| Column | Found | Expected |
+----------------+---------+----------+
| CRSElapsedTime | float64 | int64 |
| TailNum | object | float64 |
+----------------+---------+----------+
The following columns also raised exceptions on conversion:
- TailNum
ValueError("could not convert string to float: 'N54711'")
Usually this is due to dask's dtype inference failing, and
*may* be fixed by specifying dtypes manually by adding:
dtype={'CRSElapsedTime': 'float64',
'TailNum': 'object'}
to the call to `read_csv`/`read_table`.
What just happened?¶
Unlike pandas.read_csv which reads in the entire file before inferring datatypes, dask.dataframe.read_csv only reads in a sample from the beginning of the file (or first file if using a glob). These inferred datatypes are then enforced when reading all partitions.
In this case, the datatypes inferred in the sample are incorrect. The first n rows have no value for CRSElapsedTime (which pandas infers as a float), and later on turn out to be strings (object dtype). Note that Dask gives an informative error message about the mismatch. When this happens you have a few options:
Specify dtypes directly using the
dtypekeyword. This is the recommended solution, as it’s the least error prone (better to be explicit than implicit) and also the most performant.Increase the size of the
samplekeyword (in bytes)Use
assume_missingto makedaskassume that columns inferred to beint(which don’t allow missing values) are actually floats (which do allow missing values). In our particular case this doesn’t apply.
In our case we’ll use the first option and directly specify the dtypes of the offending columns.
[10]:
df = dd.read_csv(os.path.join('data', 'nycflights', '*.csv'),
parse_dates={'Date': [0, 1, 2]},
dtype={'TailNum': str,
'CRSElapsedTime': float,
'Cancelled': bool})
[11]:
df.tail() # now works
[11]:
| Date | DayOfWeek | DepTime | CRSDepTime | ArrTime | CRSArrTime | UniqueCarrier | FlightNum | TailNum | ActualElapsedTime | ... | AirTime | ArrDelay | DepDelay | Origin | Dest | Distance | TaxiIn | TaxiOut | Cancelled | Diverted | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 994 | 1999-01-25 | 1 | 632.0 | 635 | 803.0 | 817 | CO | 437 | N27213 | 91.0 | ... | 68.0 | -14.0 | -3.0 | EWR | RDU | 416.0 | 4.0 | 19.0 | False | 0 |
| 995 | 1999-01-26 | 2 | 632.0 | 635 | 751.0 | 817 | CO | 437 | N16217 | 79.0 | ... | 62.0 | -26.0 | -3.0 | EWR | RDU | 416.0 | 3.0 | 14.0 | False | 0 |
| 996 | 1999-01-27 | 3 | 631.0 | 635 | 756.0 | 817 | CO | 437 | N12216 | 85.0 | ... | 66.0 | -21.0 | -4.0 | EWR | RDU | 416.0 | 4.0 | 15.0 | False | 0 |
| 997 | 1999-01-28 | 4 | 629.0 | 635 | 803.0 | 817 | CO | 437 | N26210 | 94.0 | ... | 69.0 | -14.0 | -6.0 | EWR | RDU | 416.0 | 5.0 | 20.0 | False | 0 |
| 998 | 1999-01-29 | 5 | 632.0 | 635 | 802.0 | 817 | CO | 437 | N12225 | 90.0 | ... | 67.0 | -15.0 | -3.0 | EWR | RDU | 416.0 | 5.0 | 18.0 | False | 0 |
5 rows × 21 columns
Computations with dask.dataframe¶
We compute the maximum of the DepDelay column. With just pandas, we would loop over each file to find the individual maximums, then find the final maximum over all the individual maximums
maxes = []
for fn in filenames:
df = pd.read_csv(fn)
maxes.append(df.DepDelay.max())
final_max = max(maxes)
We could wrap that pd.read_csv with dask.delayed so that it runs in parallel. Regardless, we’re still having to think about loops, intermediate results (one per file) and the final reduction (max of the intermediate maxes). This is just noise around the real task, which pandas solves with
df = pd.read_csv(filename, dtype=dtype)
df.DepDelay.max()
dask.dataframe lets us write pandas-like code, that operates on larger than memory datasets in parallel.
[12]:
%time df.DepDelay.max().compute()
CPU times: user 51.2 ms, sys: 0 ns, total: 51.2 ms
Wall time: 531 ms
[12]:
409.0
This writes the delayed computation for us and then runs it.
Some things to note:
As with
dask.delayed, we need to call.compute()when we’re done. Up until this point everything is lazy.Dask will delete intermediate results (like the full pandas dataframe for each file) as soon as possible.
This lets us handle datasets that are larger than memory
This means that repeated computations will have to load all of the data in each time (run the code above again, is it faster or slower than you would expect?)
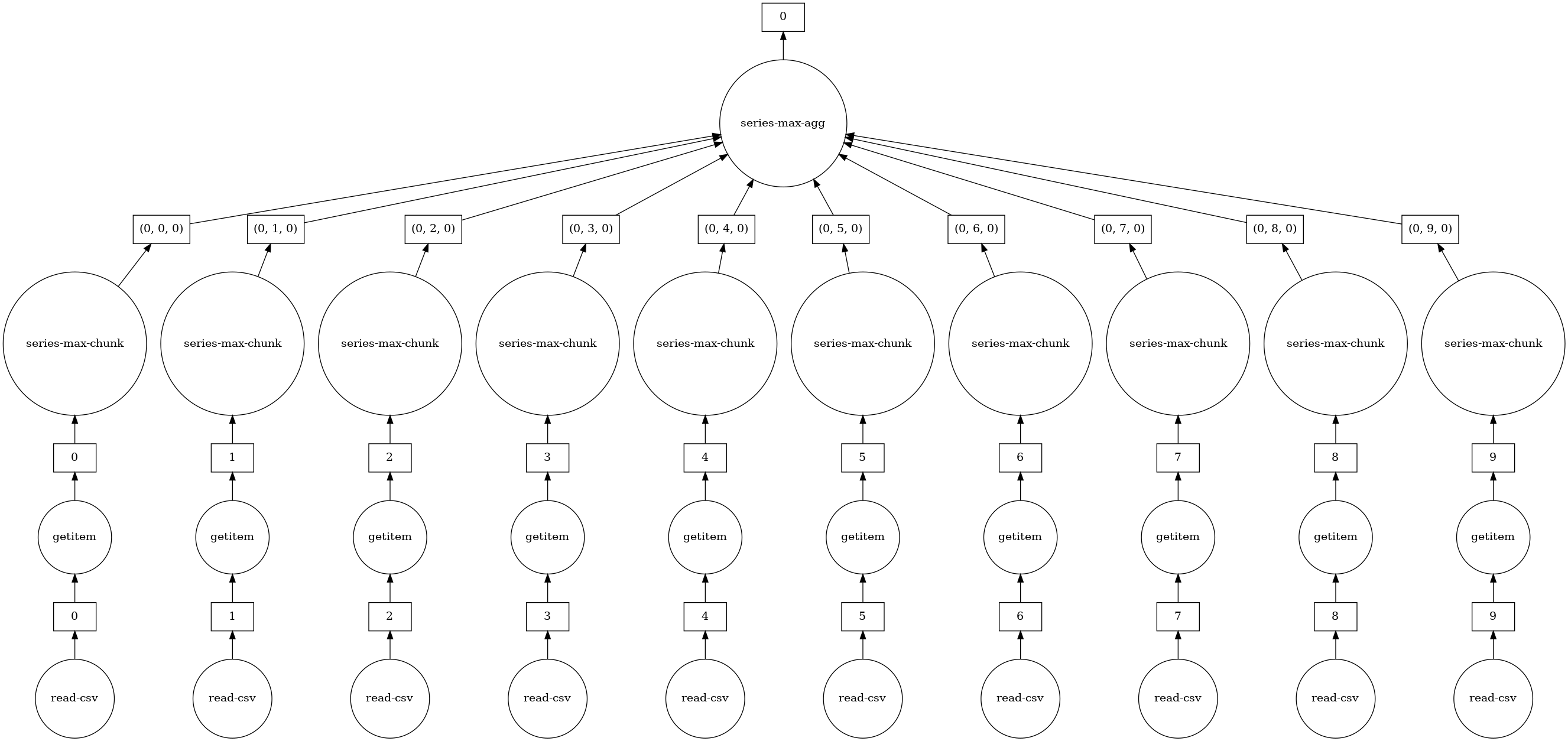
As with Delayed objects, you can view the underlying task graph using the .visualize method:
[13]:
# notice the parallelism
df.DepDelay.max().visualize()
[13]:
Exercises¶
In this section we do a few dask.dataframe computations. If you are comfortable with Pandas then these should be familiar. You will have to think about when to call compute.
1.) How many rows are in our dataset?¶
If you aren’t familiar with pandas, how would you check how many records are in a list of tuples?
[14]:
# Your code here
[15]:
len(df)
[15]:
9990
2.) In total, how many non-canceled flights were taken?¶
With pandas, you would use boolean indexing.
[16]:
# Your code here
[17]:
len(df[~df.Cancelled])
[17]:
9383
3.) In total, how many non-cancelled flights were taken from each airport?¶
Hint: use `df.groupby <https://pandas.pydata.org/pandas-docs/stable/groupby.html>`__.
[18]:
# Your code here
[19]:
df[~df.Cancelled].groupby('Origin').Origin.count().compute()
[19]:
Origin
EWR 4132
JFK 1085
LGA 4166
Name: Origin, dtype: int64
4.) What was the average departure delay from each airport?¶
Note, this is the same computation you did in the previous notebook (is this approach faster or slower?)
[20]:
# Your code here
[21]:
df.groupby("Origin").DepDelay.mean().compute()
[21]:
Origin
EWR 12.500968
JFK 17.053456
LGA 10.169227
Name: DepDelay, dtype: float64
5.) What day of the week has the worst average departure delay?¶
[22]:
# Your code here
[23]:
df.groupby("DayOfWeek").DepDelay.mean().compute()
[23]:
DayOfWeek
1 10.677698
2 8.633310
3 14.208160
4 14.187853
5 15.209929
6 9.540307
7 10.609375
Name: DepDelay, dtype: float64
Sharing Intermediate Results¶
When computing all of the above, we sometimes did the same operation more than once. For most operations, dask.dataframe hashes the arguments, allowing duplicate computations to be shared, and only computed once.
For example, lets compute the mean and standard deviation for departure delay of all non-canceled flights. Since dask operations are lazy, those values aren’t the final results yet. They’re just the recipe required to get the result.
If we compute them with two calls to compute, there is no sharing of intermediate computations.
[24]:
non_cancelled = df[~df.Cancelled]
mean_delay = non_cancelled.DepDelay.mean()
std_delay = non_cancelled.DepDelay.std()
[25]:
%%time
mean_delay_res = mean_delay.compute()
std_delay_res = std_delay.compute()
CPU times: user 79.6 ms, sys: 10.4 ms, total: 90 ms
Wall time: 315 ms
But let’s try by passing both to a single compute call.
[26]:
%%time
mean_delay_res, std_delay_res = dask.compute(mean_delay, std_delay)
CPU times: user 56.7 ms, sys: 50 µs, total: 56.8 ms
Wall time: 185 ms
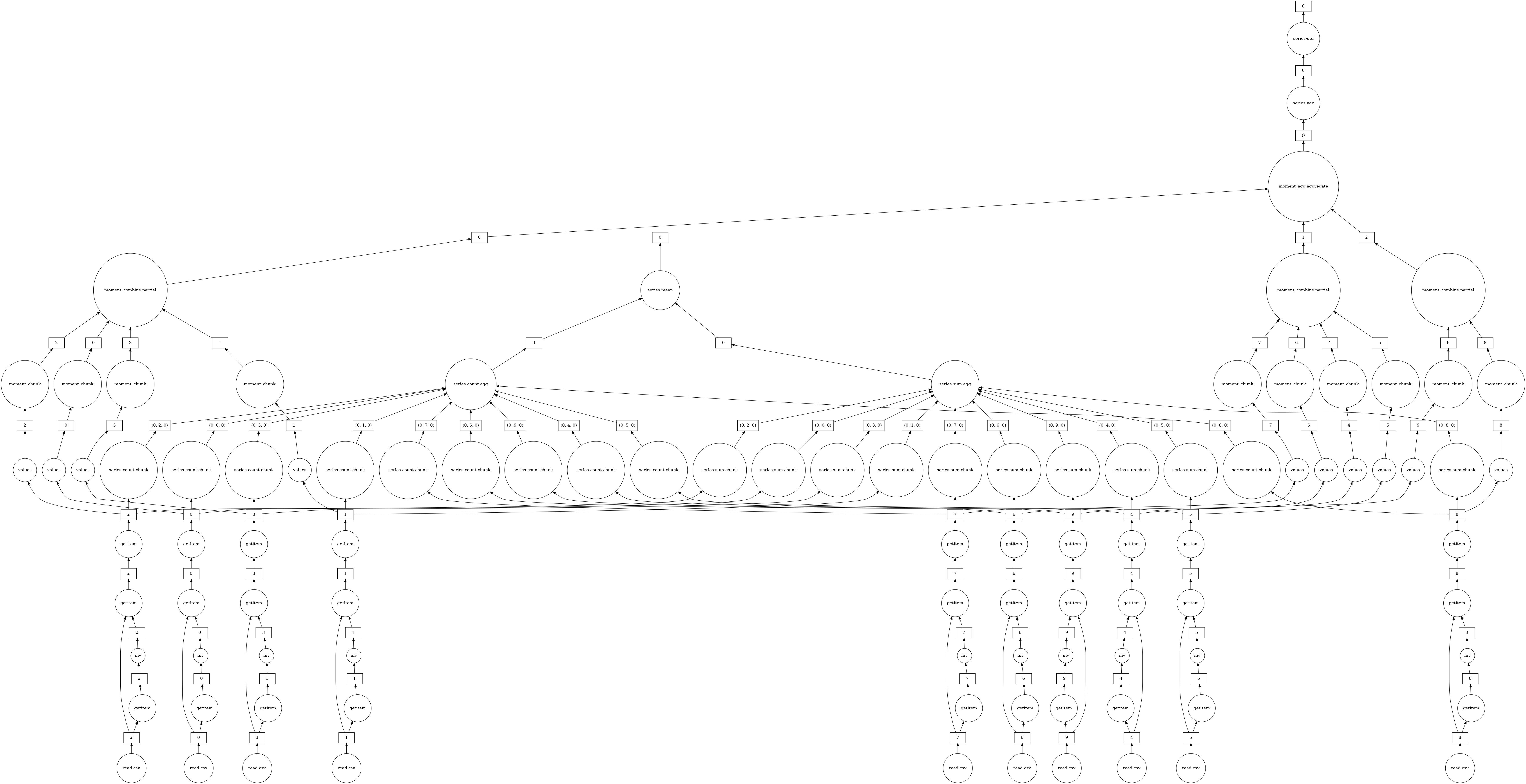
Using dask.compute takes roughly 1/2 the time. This is because the task graphs for both results are merged when calling dask.compute, allowing shared operations to only be done once instead of twice. In particular, using dask.compute only does the following once:
the calls to
read_csvthe filter (
df[~df.Cancelled])some of the necessary reductions (
sum,count)
To see what the merged task graphs between multiple results look like (and what’s shared), you can use the dask.visualize function (we might want to use filename='graph.pdf' to save the graph to disk so that we can zoom in more easily):
[27]:
dask.visualize(mean_delay, std_delay)
[27]:
How does this compare to Pandas?¶
Pandas is more mature and fully featured than dask.dataframe. If your data fits in memory then you should use Pandas. The dask.dataframe module gives you a limited pandas experience when you operate on datasets that don’t fit comfortably in memory.
During this tutorial we provide a small dataset consisting of a few CSV files. This dataset is 45MB on disk that expands to about 400MB in memory. This dataset is small enough that you would normally use Pandas.
We’ve chosen this size so that exercises finish quickly. Dask.dataframe only really becomes meaningful for problems significantly larger than this, when Pandas breaks with the dreaded
MemoryError: ...
Furthermore, the distributed scheduler allows the same dataframe expressions to be executed across a cluster. To enable massive “big data” processing, one could execute data ingestion functions such as read_csv, where the data is held on storage accessible to every worker node (e.g., amazon’s S3), and because most operations begin by selecting only some columns, transforming and filtering the data, only relatively small amounts of data need to be communicated between the machines.
Dask.dataframe operations use pandas operations internally. Generally they run at about the same speed except in the following two cases:
Dask introduces a bit of overhead, around 1ms per task. This is usually negligible.
When Pandas releases the GIL
dask.dataframecan call several pandas operations in parallel within a process, increasing speed somewhat proportional to the number of cores. For operations which don’t release the GIL, multiple processes would be needed to get the same speedup.
Dask DataFrame Data Model¶
For the most part, a Dask DataFrame feels like a pandas DataFrame. So far, the biggest difference we’ve seen is that Dask operations are lazy; they build up a task graph instead of executing immediately (more details coming in Schedulers). This lets Dask do operations in parallel and out of core.
In Dask Arrays, we saw that a dask.array was composed of many NumPy arrays, chunked along one or more dimensions. It’s similar for dask.dataframe: a Dask DataFrame is composed of many pandas DataFrames. For dask.dataframe the chunking happens only along the index.
We call each chunk a partition, and the upper / lower bounds are divisions. Dask can store information about the divisions. For now, partitions come up when you write custom functions to apply to Dask DataFrames
Converting CRSDepTime to a timestamp¶
This dataset stores timestamps as HHMM, which are read in as integers in read_csv:
[28]:
crs_dep_time = df.CRSDepTime.head(10)
crs_dep_time
[28]:
0 1540
1 1540
2 1540
3 1540
4 1540
5 1540
6 1540
7 1540
8 1540
9 1540
Name: CRSDepTime, dtype: int64
To convert these to timestamps of scheduled departure time, we need to convert these integers into pd.Timedelta objects, and then combine them with the Date column.
In pandas we’d do this using the pd.to_timedelta function, and a bit of arithmetic:
[29]:
import pandas as pd
# Get the first 10 dates to complement our `crs_dep_time`
date = df.Date.head(10)
# Get hours as an integer, convert to a timedelta
hours = crs_dep_time // 100
hours_timedelta = pd.to_timedelta(hours, unit='h')
# Get minutes as an integer, convert to a timedelta
minutes = crs_dep_time % 100
minutes_timedelta = pd.to_timedelta(minutes, unit='m')
# Apply the timedeltas to offset the dates by the departure time
departure_timestamp = date + hours_timedelta + minutes_timedelta
departure_timestamp
[29]:
0 1990-01-01 15:40:00
1 1990-01-02 15:40:00
2 1990-01-03 15:40:00
3 1990-01-04 15:40:00
4 1990-01-05 15:40:00
5 1990-01-06 15:40:00
6 1990-01-07 15:40:00
7 1990-01-08 15:40:00
8 1990-01-09 15:40:00
9 1990-01-10 15:40:00
dtype: datetime64[ns]
Custom code and Dask Dataframe¶
We could swap out pd.to_timedelta for dd.to_timedelta and do the same operations on the entire dask DataFrame. But let’s say that Dask hadn’t implemented a dd.to_timedelta that works on Dask DataFrames. What would you do then?
dask.dataframe provides a few methods to make applying custom functions to Dask DataFrames easier:
`map_partitions<http://dask.pydata.org/en/latest/dataframe-api.html#dask.dataframe.DataFrame.map_partitions>`__`map_overlap<http://dask.pydata.org/en/latest/dataframe-api.html#dask.dataframe.DataFrame.map_overlap>`__`reduction<http://dask.pydata.org/en/latest/dataframe-api.html#dask.dataframe.DataFrame.reduction>`__
Here we’ll just be discussing map_partitions, which we can use to implement to_timedelta on our own:
[30]:
# Look at the docs for `map_partitions`
help(df.CRSDepTime.map_partitions)
Help on method map_partitions in module dask.dataframe.core:
map_partitions(func, *args, **kwargs) method of dask.dataframe.core.Series instance
Apply Python function on each DataFrame partition.
Note that the index and divisions are assumed to remain unchanged.
Parameters
----------
func : function
Function applied to each partition.
args, kwargs :
Arguments and keywords to pass to the function. The partition will
be the first argument, and these will be passed *after*. Arguments
and keywords may contain ``Scalar``, ``Delayed`` or regular
python objects. DataFrame-like args (both dask and pandas) will be
repartitioned to align (if necessary) before applying the function.
meta : pd.DataFrame, pd.Series, dict, iterable, tuple, optional
An empty ``pd.DataFrame`` or ``pd.Series`` that matches the dtypes
and column names of the output. This metadata is necessary for
many algorithms in dask dataframe to work. For ease of use, some
alternative inputs are also available. Instead of a ``DataFrame``,
a ``dict`` of ``{name: dtype}`` or iterable of ``(name, dtype)``
can be provided (note that the order of the names should match the
order of the columns). Instead of a series, a tuple of ``(name,
dtype)`` can be used. If not provided, dask will try to infer the
metadata. This may lead to unexpected results, so providing
``meta`` is recommended. For more information, see
``dask.dataframe.utils.make_meta``.
Examples
--------
Given a DataFrame, Series, or Index, such as:
>>> import dask.dataframe as dd
>>> df = pd.DataFrame({'x': [1, 2, 3, 4, 5],
... 'y': [1., 2., 3., 4., 5.]})
>>> ddf = dd.from_pandas(df, npartitions=2)
One can use ``map_partitions`` to apply a function on each partition.
Extra arguments and keywords can optionally be provided, and will be
passed to the function after the partition.
Here we apply a function with arguments and keywords to a DataFrame,
resulting in a Series:
>>> def myadd(df, a, b=1):
... return df.x + df.y + a + b
>>> res = ddf.map_partitions(myadd, 1, b=2)
>>> res.dtype
dtype('float64')
By default, dask tries to infer the output metadata by running your
provided function on some fake data. This works well in many cases, but
can sometimes be expensive, or even fail. To avoid this, you can
manually specify the output metadata with the ``meta`` keyword. This
can be specified in many forms, for more information see
``dask.dataframe.utils.make_meta``.
Here we specify the output is a Series with no name, and dtype
``float64``:
>>> res = ddf.map_partitions(myadd, 1, b=2, meta=(None, 'f8'))
Here we map a function that takes in a DataFrame, and returns a
DataFrame with a new column:
>>> res = ddf.map_partitions(lambda df: df.assign(z=df.x * df.y))
>>> res.dtypes
x int64
y float64
z float64
dtype: object
As before, the output metadata can also be specified manually. This
time we pass in a ``dict``, as the output is a DataFrame:
>>> res = ddf.map_partitions(lambda df: df.assign(z=df.x * df.y),
... meta={'x': 'i8', 'y': 'f8', 'z': 'f8'})
In the case where the metadata doesn't change, you can also pass in
the object itself directly:
>>> res = ddf.map_partitions(lambda df: df.head(), meta=ddf)
Also note that the index and divisions are assumed to remain unchanged.
If the function you're mapping changes the index/divisions, you'll need
to clear them afterwards:
>>> ddf.map_partitions(func).clear_divisions() # doctest: +SKIP
The basic idea is to apply a function that operates on a DataFrame to each partition. In this case, we’ll apply pd.to_timedelta.
[31]:
hours = df.CRSDepTime // 100
# hours_timedelta = pd.to_timedelta(hours, unit='h')
hours_timedelta = hours.map_partitions(pd.to_timedelta, unit='h')
minutes = df.CRSDepTime % 100
# minutes_timedelta = pd.to_timedelta(minutes, unit='m')
minutes_timedelta = minutes.map_partitions(pd.to_timedelta, unit='m')
departure_timestamp = df.Date + hours_timedelta + minutes_timedelta
[32]:
departure_timestamp
[32]:
Dask Series Structure:
npartitions=10
datetime64[ns]
...
...
...
...
dtype: datetime64[ns]
Dask Name: add, 90 tasks
[33]:
departure_timestamp.head()
[33]:
0 1990-01-01 15:40:00
1 1990-01-02 15:40:00
2 1990-01-03 15:40:00
3 1990-01-04 15:40:00
4 1990-01-05 15:40:00
dtype: datetime64[ns]
Exercise: Rewrite above to use a single call to map_partitions¶
This will be slightly more efficient than two separate calls, as it reduces the number of tasks in the graph.
[34]:
def compute_departure_timestamp(df):
pass # TODO: implement this
[35]:
departure_timestamp = df.map_partitions(compute_departure_timestamp)
departure_timestamp.head()
distributed.worker - WARNING - Compute Failed
Function: execute_task
args: ((<function safe_head at 0x7f9cb91c2c10>, (subgraph_callable, (<function pandas_read_text at 0x7f9cb9177280>, <function read_csv at 0x7f9cba4e5700>, (<function read_block_from_file at 0x7f9cb9107160>, <OpenFile '/home/runner/work/dask-tutorial/dask-tutorial/data/nycflights/1990.csv'>, 0, 64000000, b'\n'), b'Year,Month,DayofMonth,DayOfWeek,DepTime,CRSDepTime,ArrTime,CRSArrTime,UniqueCarrier,FlightNum,TailNum,ActualElapsedTime,CRSElapsedTime,AirTime,ArrDelay,DepDelay,Origin,Dest,Distance,TaxiIn,TaxiOut,Cancelled,Diverted\n', {'parse_dates': {'Date': [0, 1, 2]}, 'dtype': {'TailNum': <class 'str'>, 'CRSElapsedTime': <class 'float'>, 'Cancelled': <class 'bool'>}}, {'Date': dtype('<M8[ns]'), 'DayOfWeek': dtype('int64'), 'DepTime': dtype('float64'), 'CRSDepTime': dtype('int64'), 'ArrTime': dtype('float64'), 'CRSArrTime': dtype('int64'), 'UniqueCarrier': dtype('O'), 'FlightNum': dtype('int64'), 'TailNum': dtype('O'), 'ActualElapsedTime': dtype('float64'), 'CRSElapsedTime': dtype('float64'), 'A
kwargs: {}
Exception: AttributeError("'NoneType' object has no attribute 'head'")
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
/tmp/ipykernel_7074/1135339802.py in <module>
1 departure_timestamp = df.map_partitions(compute_departure_timestamp)
2
----> 3 departure_timestamp.head()
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/dask/dataframe/core.py in head(self, n, npartitions, compute)
1003 Whether to compute the result, default is True.
1004 """
-> 1005 return self._head(n=n, npartitions=npartitions, compute=compute, safe=True)
1006
1007 def _head(self, n, npartitions, compute, safe):
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/dask/dataframe/core.py in _head(self, n, npartitions, compute, safe)
1036
1037 if compute:
-> 1038 result = result.compute()
1039 return result
1040
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/dask/base.py in compute(self, **kwargs)
164 dask.base.compute
165 """
--> 166 (result,) = compute(self, traverse=False, **kwargs)
167 return result
168
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/dask/base.py in compute(*args, **kwargs)
442 postcomputes.append(x.__dask_postcompute__())
443
--> 444 results = schedule(dsk, keys, **kwargs)
445 return repack([f(r, *a) for r, (f, a) in zip(results, postcomputes)])
446
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/distributed/client.py in get(self, dsk, keys, restrictions, loose_restrictions, resources, sync, asynchronous, direct, retries, priority, fifo_timeout, actors, **kwargs)
2680 should_rejoin = False
2681 try:
-> 2682 results = self.gather(packed, asynchronous=asynchronous, direct=direct)
2683 finally:
2684 for f in futures.values():
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/distributed/client.py in gather(self, futures, errors, direct, asynchronous)
1974 else:
1975 local_worker = None
-> 1976 return self.sync(
1977 self._gather,
1978 futures,
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/distributed/client.py in sync(self, func, asynchronous, callback_timeout, *args, **kwargs)
829 return future
830 else:
--> 831 return sync(
832 self.loop, func, *args, callback_timeout=callback_timeout, **kwargs
833 )
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/distributed/utils.py in sync(loop, func, callback_timeout, *args, **kwargs)
337 if error[0]:
338 typ, exc, tb = error[0]
--> 339 raise exc.with_traceback(tb)
340 else:
341 return result[0]
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/distributed/utils.py in f()
321 if callback_timeout is not None:
322 future = asyncio.wait_for(future, callback_timeout)
--> 323 result[0] = yield future
324 except Exception as exc:
325 error[0] = sys.exc_info()
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/tornado/gen.py in run(self)
760
761 try:
--> 762 value = future.result()
763 except Exception:
764 exc_info = sys.exc_info()
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/distributed/client.py in _gather(self, futures, errors, direct, local_worker)
1839 exc = CancelledError(key)
1840 else:
-> 1841 raise exception.with_traceback(traceback)
1842 raise exc
1843 if errors == "skip":
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/dask/dataframe/core.py in safe_head()
6169
6170 def safe_head(df, n):
-> 6171 r = M.head(df, n)
6172 if len(r) != n:
6173 msg = (
/usr/share/miniconda3/envs/dask-tutorial/lib/python3.8/site-packages/dask/utils.py in __call__()
893
894 def __call__(self, obj, *args, **kwargs):
--> 895 return getattr(obj, self.method)(*args, **kwargs)
896
897 def __reduce__(self):
AttributeError: 'NoneType' object has no attribute 'head'
[36]:
def compute_departure_timestamp(df):
hours = df.CRSDepTime // 100
hours_timedelta = pd.to_timedelta(hours, unit='h')
minutes = df.CRSDepTime % 100
minutes_timedelta = pd.to_timedelta(minutes, unit='m')
return df.Date + hours_timedelta + minutes_timedelta
departure_timestamp = df.map_partitions(compute_departure_timestamp)
departure_timestamp.head()
[36]:
0 1990-01-01 15:40:00
1 1990-01-02 15:40:00
2 1990-01-03 15:40:00
3 1990-01-04 15:40:00
4 1990-01-05 15:40:00
dtype: datetime64[ns]
Limitations¶
What doesn’t work?¶
Dask.dataframe only covers a small but well-used portion of the Pandas API. This limitation is for two reasons:
The Pandas API is huge
Some operations are genuinely hard to do in parallel (e.g. sort)
Additionally, some important operations like set_index work, but are slower than in Pandas because they include substantial shuffling of data, and may write out to disk.
Learn More¶
[37]:
client.shutdown()